Encontrarás explicación sobre selección de modelo en regresión con más de dos variables independientes en páginas 226 – 231, Experimentación en Agricultura.
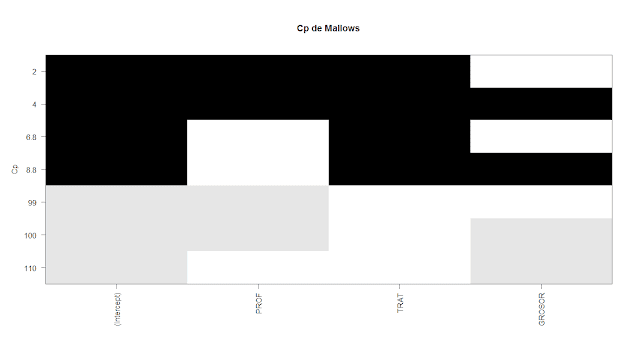 |
plot(todos, scale = "Cp" , main = "Cp de Mallows")
|
El Cp con
el menor valor y el R2-ajustado con el mayor, sirven para
seleccionar el modelo con las variables más apropiadas.
Códigos para el software R
SUELO <- read.csv("C:/Users/Administrator/Desktop/tabla 16.2.csv", header=TRUE)
attach(SUELO); knitr::kable(SUELO)
| 26.34 | 0 | 0.5 | 20 |
| 28.25 | 0 | 0.5 | 30 |
| 26.24 | 0 | 0.5 | 50 |
| 37.28 | 0 | 0.5 | 70 |
| 42.04 | 0 | 0.5 | 90 |
| 21.15 | 0 | 1.0 | 20 |
| 34.38 | 0 | 1.0 | 30 |
| 26.67 | 0 | 1.0 | 50 |
| 28.39 | 0 | 1.0 | 70 |
| 40.50 | 0 | 1.0 | 90 |
| 35.53 | 0 | 2.0 | 20 |
| 22.30 | 0 | 2.0 | 30 |
| 20.75 | 0 | 2.0 | 50 |
| 20.35 | 0 | 2.0 | 70 |
| 47.32 | 0 | 2.0 | 90 |
| 44.75 | 1 | 0.5 | 20 |
| 74.16 | 1 | 0.5 | 30 |
| 62.11 | 1 | 0.5 | 50 |
| 57.41 | 1 | 0.5 | 70 |
| 60.78 | 1 | 0.5 | 90 |
| 41.54 | 1 | 1.0 | 20 |
| 61.10 | 1 | 1.0 | 30 |
| 63.67 | 1 | 1.0 | 50 |
| 62.31 | 1 | 1.0 | 70 |
| 65.21 | 1 | 1.0 | 90 |
| 51.80 | 1 | 2.0 | 20 |
| 74.04 | 1 | 2.0 | 30 |
| 60.69 | 1 | 2.0 | 50 |
| 56.52 | 1 | 2.0 | 70 |
| 69.23 | 1 | 2.0 | 90 |
library(leaps)
library(HH)
todos <- regsubsets(N_MINERAL ~ PROF + TRAT + GROSOR, data = SUELO, nbest=10)
summary(todos); stars <- summary(todos); as.data.frame(stars$outmat)
## Subset selection object
## Call: regsubsets.formula(N_MINERAL ~ PROF + TRAT + GROSOR, data = SUELO,
## nbest = 10)
## 3 Variables (and intercept)
## Forced in Forced out
## PROF FALSE FALSE
## TRAT FALSE FALSE
## GROSOR FALSE FALSE
## 10 subsets of each size up to 3
## Selection Algorithm: exhaustive
## PROF TRAT GROSOR
## 1 ( 1 ) " " "*" " "
## 1 ( 2 ) "*" " " " "
## 1 ( 3 ) " " " " "*"
## 2 ( 1 ) "*" "*" " "
## 2 ( 2 ) " " "*" "*"
## 2 ( 3 ) "*" " " "*"
## 3 ( 1 ) "*" "*" "*"
## PROF TRAT GROSOR
## 1 ( 1 ) *
## 1 ( 2 ) *
## 1 ( 3 ) *
## 2 ( 1 ) * *
## 2 ( 2 ) * *
## 2 ( 3 ) * *
## 3 ( 1 ) * * *
knitr::kable(summaryHH(todos))
| T | 2 | 0.7517322 | 2207.813 | 0.7428655 | 6.825148 | -34.995022 | 8.879779 |
| P | 2 | 0.0515926 | 8434.064 | 0.0177209 | 99.395289 | 5.213262 | 17.355592 |
| G | 2 | 0.0000283 | 8892.618 | -0.0356849 | 106.212940 | 6.801544 | 17.821154 |
| P-T | 3 | 0.8033248 | 1749.007 | 0.7887562 | 2.003748 | -38.582452 | 8.048481 |
| T-G | 3 | 0.7517605 | 2207.561 | 0.7333724 | 8.821400 | -31.597251 | 9.042208 |
| P-G | 3 | 0.0516209 | 8433.812 | -0.0186294 | 101.391541 | 8.613563 | 17.673806 |
| P-T-G | 4 | 0.8033531 | 1748.755 | 0.7806631 | 4.000000 | -35.185579 | 8.201208 |
knitr::kable(data.frame(summary(todos)$cp, summary(todos)$adjr2, summary(todos)$rsq, summary(todos)$rss))
| 6.825148 | 0.7428655 | 0.7517322 | 2207.813 |
| 99.395289 | 0.0177209 | 0.0515926 | 8434.064 |
| 106.212940 | -0.0356849 | 0.0000283 | 8892.618 |
| 2.003748 | 0.7887562 | 0.8033248 | 1749.007 |
| 8.821400 | 0.7333724 | 0.7517605 | 2207.561 |
| 101.391541 | -0.0186294 | 0.0516209 | 8433.812 |
| 4.000000 | 0.7806631 | 0.8033531 | 1748.755 |
plot(todos, scale = "r2" , main = "Coeficiente de Determinación R^2")
plot(todos, scale = "Cp" , main = "Cp de Mallows")
plot(todos, scale = "adjr2", main = "R^2 ajustado")
plot(todos, scale = "bic" , main = "Criterio de información bayesiano (BIC)")
modelo.completo <- lm(N_MINERAL ~ PROF + TRAT + GROSOR, data = SUELO)
modelo.reducido <- step(modelo.completo, direction = "backward")
## Start: AIC=129.96
## N_MINERAL ~ PROF + TRAT + GROSOR
##
## Df Sum of Sq RSS AIC
## - GROSOR 1 0.3 1749.0 127.97
## <none> 1748.8 129.96
## - PROF 1 458.8 2207.6 134.95
## - TRAT 1 6685.1 8433.8 175.16
##
## Step: AIC=127.97
## N_MINERAL ~ PROF + TRAT
##
## Df Sum of Sq RSS AIC
## <none> 1749.0 127.97
## - PROF 1 458.8 2207.8 132.96
## - TRAT 1 6685.1 8434.1 173.16
summary(modelo.reducido); modelo.reducido$anova
##
## Call:
## lm(formula = N_MINERAL ~ PROF + TRAT, data = SUELO)
##
## Residuals:
## Min 1Q Median 3Q Max
## -13.9287 -4.8533 -0.0762 4.0864 17.1644
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 22.55961 3.63579 6.205 1.24e-06 ***
## PROF 0.15269 0.05737 2.661 0.0129 *
## TRAT 29.85533 2.93889 10.159 1.01e-10 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 8.048 on 27 degrees of freedom
## Multiple R-squared: 0.8033, Adjusted R-squared: 0.7888
## F-statistic: 55.14 on 2 and 27 DF, p-value: 2.922e-10
## Step Df Deviance Resid. Df Resid. Dev AIC
## 1 NA NA 26 1748.755 129.9639
## 2 - GROSOR 1 0.252105 27 1749.007 127.9682
modelo.fwd <- step(lm(N_MINERAL ~1), direction = "forward", scope = (~PROF + TRAT + GROSOR))
## Start: AIC=172.75
## N_MINERAL ~ 1
##
## Df Sum of Sq RSS AIC
## + TRAT 1 6685.1 2207.8 132.96
## <none> 8892.9 172.75
## + PROF 1 458.8 8434.1 173.16
## + GROSOR 1 0.3 8892.6 174.75
##
## Step: AIC=132.96
## N_MINERAL ~ TRAT
##
## Df Sum of Sq RSS AIC
## + PROF 1 458.81 1749.0 127.97
## <none> 2207.8 132.96
## + GROSOR 1 0.25 2207.6 134.95
##
## Step: AIC=127.97
## N_MINERAL ~ TRAT + PROF
##
## Df Sum of Sq RSS AIC
## <none> 1749.0 127.97
## + GROSOR 1 0.25211 1748.8 129.96
summary(modelo.fwd); modelo.fwd$anova
##
## Call:
## lm(formula = N_MINERAL ~ TRAT + PROF)
##
## Residuals:
## Min 1Q Median 3Q Max
## -13.9287 -4.8533 -0.0762 4.0864 17.1644
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 22.55961 3.63579 6.205 1.24e-06 ***
## TRAT 29.85533 2.93889 10.159 1.01e-10 ***
## PROF 0.15269 0.05737 2.661 0.0129 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 8.048 on 27 degrees of freedom
## Multiple R-squared: 0.8033, Adjusted R-squared: 0.7888
## F-statistic: 55.14 on 2 and 27 DF, p-value: 2.922e-10
## Step Df Deviance Resid. Df Resid. Dev AIC
## 1 NA NA 29 8892.870 172.7542
## 2 + TRAT -1 6685.0570 28 2207.813 132.9568
## 3 + PROF -1 458.8061 27 1749.007 127.9682
modelo.completo <- lm(N_MINERAL ~., data=SUELO)
MSE=(summary(modelo.completo)$sigma)^2
intercepto <- lm(N_MINERAL~1)
modelos <- step(intercepto, scope = list(upper = modelo.completo), scale = MSE); modelos
## Start: AIC=104.22
## N_MINERAL ~ 1
##
## Df Sum of Sq RSS Cp
## + TRAT 1 6685.1 2207.8 6.8251
## + PROF 1 458.8 8434.1 99.3953
## <none> 8892.9 104.2167
## + GROSOR 1 0.3 8892.6 106.2129
##
## Step: AIC=6.83
## N_MINERAL ~ TRAT
##
## Df Sum of Sq RSS Cp
## + PROF 1 458.8 1749.0 2.0037
## <none> 2207.8 6.8251
## + GROSOR 1 0.3 2207.6 8.8214
## - TRAT 1 6685.1 8892.9 104.2167
##
## Step: AIC=2
## N_MINERAL ~ TRAT + PROF
##
## Df Sum of Sq RSS Cp
## <none> 1749.0 2.0037
## + GROSOR 1 0.3 1748.8 4.0000
## - PROF 1 458.8 2207.8 6.8251
## - TRAT 1 6685.1 8434.1 99.3953
##
## Call:
## lm(formula = N_MINERAL ~ TRAT + PROF)
##
## Coefficients:
## (Intercept) TRAT PROF
## 22.5596 29.8553 0.1527
summary(modelos); modelos$anova
##
## Call:
## lm(formula = N_MINERAL ~ TRAT + PROF)
##
## Residuals:
## Min 1Q Median 3Q Max
## -13.9287 -4.8533 -0.0762 4.0864 17.1644
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 22.55961 3.63579 6.205 1.24e-06 ***
## TRAT 29.85533 2.93889 10.159 1.01e-10 ***
## PROF 0.15269 0.05737 2.661 0.0129 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 8.048 on 27 degrees of freedom
## Multiple R-squared: 0.8033, Adjusted R-squared: 0.7888
## F-statistic: 55.14 on 2 and 27 DF, p-value: 2.922e-10
## Step Df Deviance Resid. Df Resid. Dev Cp
## 1 NA NA 29 8892.870 104.216688
## 2 + TRAT -1 6685.0570 28 2207.813 6.825148
## 3 + PROF -1 458.8061 27 1749.007 2.003748
modelo.final <- lm(N_MINERAL ~ PROF + TRAT, data = SUELO)
anova(modelo.final); summary(modelo.final)
## Analysis of Variance Table
##
## Response: N_MINERAL
## Df Sum Sq Mean Sq F value Pr(>F)
## PROF 1 458.8 458.8 7.0827 0.01294 *
## TRAT 1 6685.1 6685.1 103.1994 1.012e-10 ***
## Residuals 27 1749.0 64.8
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Call:
## lm(formula = N_MINERAL ~ PROF + TRAT, data = SUELO)
##
## Residuals:
## Min 1Q Median 3Q Max
## -13.9287 -4.8533 -0.0762 4.0864 17.1644
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 22.55961 3.63579 6.205 1.24e-06 ***
## PROF 0.15269 0.05737 2.661 0.0129 *
## TRAT 29.85533 2.93889 10.159 1.01e-10 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 8.048 on 27 degrees of freedom
## Multiple R-squared: 0.8033, Adjusted R-squared: 0.7888
## F-statistic: 55.14 on 2 and 27 DF, p-value: 2.922e-10
library(broom)
tidy(modelo.final); glance(modelo.final)
## term estimate std.error statistic p.value
## 1 (Intercept) 22.559610 3.63578542 6.204879 1.235274e-06
## 2 PROF 0.152687 0.05737219 2.661342 1.294477e-02
## 3 TRAT 29.855333 2.93888984 10.158711 1.011703e-10
## r.squared adj.r.squared sigma statistic p.value df logLik
## 1 0.8033248 0.7887562 8.048481 55.14108 2.921599e-10 3 -103.5523
## AIC BIC deviance df.residual
## 1 215.1045 220.7093 1749.007 27
knitr::kable(augment(modelo.final))
| 26.34 | 20 | 0 | 25.61335 | 2.772923 | 0.7266504 | 0.1186992 | 8.200395 | 0.0004152 | 0.0961722 |
| 28.25 | 30 | 0 | 27.14022 | 2.431390 | 1.1097805 | 0.0912602 | 8.198621 | 0.0007004 | 0.1446450 |
| 26.24 | 50 | 0 | 30.19396 | 2.081274 | -3.9539593 | 0.0668699 | 8.162422 | 0.0061782 | -0.5085658 |
| 37.28 | 70 | 0 | 33.24770 | 2.320561 | 4.0323008 | 0.0831301 | 8.160113 | 0.0082737 | 0.5232209 |
| 42.04 | 90 | 0 | 36.30144 | 3.011903 | 5.7385610 | 0.1400407 | 8.111515 | 0.0320889 | 0.7688650 |
| 21.15 | 20 | 0 | 25.61335 | 2.772923 | -4.4633496 | 0.1186992 | 8.148626 | 0.0156665 | -0.5907244 |
| 34.38 | 30 | 0 | 27.14022 | 2.431390 | 7.2397805 | 0.0912602 | 8.065428 | 0.0298060 | 0.9436081 |
| 26.67 | 50 | 0 | 30.19396 | 2.081274 | -3.5239593 | 0.0668699 | 8.170536 | 0.0049075 | -0.4532584 |
| 28.39 | 70 | 0 | 33.24770 | 2.320561 | -4.8576992 | 0.0831301 | 8.141231 | 0.0120076 | -0.6303225 |
| 40.50 | 90 | 0 | 36.30144 | 3.011903 | 4.1985610 | 0.1400407 | 8.153595 | 0.0171771 | 0.5625324 |
| 35.53 | 20 | 0 | 25.61335 | 2.772923 | 9.9166504 | 0.1186992 | 7.935854 | 0.0773357 | 1.3124688 |
| 22.30 | 30 | 0 | 27.14022 | 2.431390 | -4.8402195 | 0.0912602 | 8.141128 | 0.0133224 | -0.6308576 |
| 20.75 | 50 | 0 | 30.19396 | 2.081274 | -9.4439593 | 0.0668699 | 7.974545 | 0.0352456 | -1.2147001 |
| 20.35 | 70 | 0 | 33.24770 | 2.320561 | -12.8976992 | 0.0831301 | 7.764748 | 0.0846483 | -1.6735722 |
| 47.32 | 90 | 0 | 36.30144 | 3.011903 | 11.0185610 | 0.1400407 | 7.863811 | 0.1183039 | 1.4762910 |
| 44.75 | 20 | 1 | 55.46868 | 2.772923 | -10.7186829 | 0.1186992 | 7.890216 | 0.0903510 | -1.4186178 |
| 74.16 | 30 | 1 | 56.99555 | 2.431390 | 17.1644472 | 0.0912602 | 7.402708 | 0.1675377 | 2.2371551 |
| 62.11 | 50 | 1 | 60.04929 | 2.081274 | 2.0607073 | 0.0668699 | 8.191122 | 0.0016781 | 0.2650521 |
| 57.41 | 70 | 1 | 63.10303 | 2.320561 | -5.6930325 | 0.0831301 | 8.118493 | 0.0164923 | -0.7387132 |
| 60.78 | 90 | 1 | 66.15677 | 3.011903 | -5.3767724 | 0.1400407 | 8.122594 | 0.0281704 | -0.7203918 |
| 41.54 | 20 | 1 | 55.46868 | 2.772923 | -13.9286829 | 0.1186992 | 7.668289 | 0.1525704 | -1.8434613 |
| 61.10 | 30 | 1 | 56.99555 | 2.431390 | 4.1044472 | 0.0912602 | 8.158217 | 0.0095799 | 0.5349596 |
| 63.67 | 50 | 1 | 60.04929 | 2.081274 | 3.6207073 | 0.0668699 | 8.168792 | 0.0051806 | 0.4657023 |
| 62.31 | 70 | 1 | 63.10303 | 2.320561 | -0.7930325 | 0.0831301 | 8.200191 | 0.0003200 | -0.1029019 |
| 65.21 | 90 | 1 | 66.15677 | 3.011903 | -0.9467724 | 0.1400407 | 8.199355 | 0.0008735 | -0.1268506 |
| 51.80 | 20 | 1 | 55.46868 | 2.772923 | -3.6686829 | 0.1186992 | 8.165913 | 0.0105845 | -0.4855502 |
| 74.04 | 30 | 1 | 56.99555 | 2.431390 | 17.0444472 | 0.0912602 | 7.414434 | 0.1652033 | 2.2215147 |
| 60.69 | 50 | 1 | 60.04929 | 2.081274 | 0.6407073 | 0.0668699 | 8.200768 | 0.0001622 | 0.0824090 |
| 56.52 | 70 | 1 | 63.10303 | 2.320561 | -6.5830325 | 0.0831301 | 8.090217 | 0.0220519 | -0.8541973 |
| 69.23 | 90 | 1 | 66.15677 | 3.011903 | 3.0732276 | 0.1400407 | 8.176008 | 0.0092032 | 0.4117578 |
intervalo <- predict(modelo.final, interval = "confidence")
prediccion <- predict(modelo.final, interval = "prediction")
knitr::kable(data.frame(N_MINERAL, intervalo, prediccion))
| 26.34 | 25.61335 | 19.92378 | 31.30292 | 25.61335 | 8.146600 | 43.08010 |
| 28.25 | 27.14022 | 22.15142 | 32.12902 | 27.14022 | 9.889009 | 44.39143 |
| 26.24 | 30.19396 | 25.92354 | 34.46438 | 30.19396 | 13.136625 | 47.25129 |
| 37.28 | 33.24770 | 28.48630 | 38.00910 | 33.24770 | 16.060871 | 50.43453 |
| 42.04 | 36.30144 | 30.12152 | 42.48135 | 36.30144 | 18.668870 | 53.93401 |
| 21.15 | 25.61335 | 19.92378 | 31.30292 | 25.61335 | 8.146600 | 43.08010 |
| 34.38 | 27.14022 | 22.15142 | 32.12902 | 27.14022 | 9.889009 | 44.39143 |
| 26.67 | 30.19396 | 25.92354 | 34.46438 | 30.19396 | 13.136625 | 47.25129 |
| 28.39 | 33.24770 | 28.48630 | 38.00910 | 33.24770 | 16.060871 | 50.43453 |
| 40.50 | 36.30144 | 30.12152 | 42.48135 | 36.30144 | 18.668870 | 53.93401 |
| 35.53 | 25.61335 | 19.92378 | 31.30292 | 25.61335 | 8.146600 | 43.08010 |
| 22.30 | 27.14022 | 22.15142 | 32.12902 | 27.14022 | 9.889009 | 44.39143 |
| 20.75 | 30.19396 | 25.92354 | 34.46438 | 30.19396 | 13.136625 | 47.25129 |
| 20.35 | 33.24770 | 28.48630 | 38.00910 | 33.24770 | 16.060871 | 50.43453 |
| 47.32 | 36.30144 | 30.12152 | 42.48135 | 36.30144 | 18.668870 | 53.93401 |
| 44.75 | 55.46868 | 49.77911 | 61.15825 | 55.46868 | 38.001934 | 72.93543 |
| 74.16 | 56.99555 | 52.00675 | 61.98435 | 56.99555 | 39.744342 | 74.24676 |
| 62.11 | 60.04929 | 55.77887 | 64.31972 | 60.04929 | 42.991959 | 77.10663 |
| 57.41 | 63.10303 | 58.34163 | 67.86443 | 63.10303 | 45.916204 | 80.28986 |
| 60.78 | 66.15677 | 59.97686 | 72.33669 | 66.15677 | 48.524203 | 83.78934 |
| 41.54 | 55.46868 | 49.77911 | 61.15825 | 55.46868 | 38.001934 | 72.93543 |
| 61.10 | 56.99555 | 52.00675 | 61.98435 | 56.99555 | 39.744342 | 74.24676 |
| 63.67 | 60.04929 | 55.77887 | 64.31972 | 60.04929 | 42.991959 | 77.10663 |
| 62.31 | 63.10303 | 58.34163 | 67.86443 | 63.10303 | 45.916204 | 80.28986 |
| 65.21 | 66.15677 | 59.97686 | 72.33669 | 66.15677 | 48.524203 | 83.78934 |
| 51.80 | 55.46868 | 49.77911 | 61.15825 | 55.46868 | 38.001934 | 72.93543 |
| 74.04 | 56.99555 | 52.00675 | 61.98435 | 56.99555 | 39.744342 | 74.24676 |
| 60.69 | 60.04929 | 55.77887 | 64.31972 | 60.04929 | 42.991959 | 77.10663 |
| 56.52 | 63.10303 | 58.34163 | 67.86443 | 63.10303 | 45.916204 | 80.28986 |
| 69.23 | 66.15677 | 59.97686 | 72.33669 | 66.15677 | 48.524203 | 83.78934 |
par(mfrow = c(1, 1))
plot(modelo.final, 1)
plot(modelo.final, 2)
plot(modelo.final, 3)
plot(modelo.final, 4)
plot(modelo.final, 5)
plot(modelo.final, 6)
Códigos para el software SAS
/***********************************************************
EXPERIMENTACIÓN EN AGRICULTURA
Fernández
Escobar, R.; Trapero, A.; Domínguez, J. 2010
CAPÍTULO 16:
REGRESIÓN MÚLTIPLE
REGRESIÓN CON
MÁS DE DOS VARIABLES INDEPENDIENTES (pp. 226-231)
************************************************************/
DATA SUELO;
INPUT N_MINERAL TRAT GROSOR PROF;
LABEL
N_MINERAL= "mg N/kg suelo"
TRAT = "kg N/árbol"
GROSOR = "mm"
PROF = "cm";
CARDS;
26.34 0 0.5 20
28.25 0 0.5 30
26.24 0 0.5 50
37.28 0 0.5 70
42.04 0 0.5 90
21.15 0 1 20
34.38 0 1 30
26.67 0 1 50
28.39 0 1 70
40.5 0 1 90
35.53 0 2 20
22.3 0 2 30
20.75 0 2 50
20.35 0 2 70
47.32 0 2 90
44.75 1 0.5 20
74.16 1 0.5 30
62.11 1 0.5 50
57.41 1 0.5 70
60.78 1 0.5 90
41.54 1 1 20
61.1 1 1 30
63.67 1 1 50
62.31 1 1 70
65.21 1 1 90
51.8 1 2 20
74.04 1 2 30
60.69 1 2 50
56.52 1 2 70
69.23 1 2 90
;
ODS HTML;
* Mostrar datos a analizar;
PROC PRINT DATA =
SUELO;
RUN;
* El primer
procedimiento: Best Subset Regression Models;
PROC REG;
MODEL
N_MINERAL = PROF TRAT GROSOR / SELECTION = CP ADJRSQ RSQUARE PRESS MSE SSE SBC SP PC B BIC AIC JP;
RUN;
* El segundo procedimiento: Stepwise Linear
Regression;
PROC REG;
MODEL
N_MINERAL = PROF TRAT GROSOR / SELECTION =
STEPWISE;
RUN;
* Regresión múltiple;
PROC REG;
MODEL
N_MINERAL = PROF TRAT / R P CLM CLI influence;
RUN;
* Correlación;
PROC CORR PEARSON;
VAR N_MINERAL PROF TRAT GROSOR; *Valor de Correlación Múltiple es
tan grande como el mayor de los coeficientes simples;
RUN;
* Correlación Parcial;
PROC CORR;
VAR
GROSOR;
WITH
N_MINERAL;
PARTIAL
PROF TRAT;
RUN;
QUIT;
ODS HTML CLOSE;
------------------------------------------------------------

























